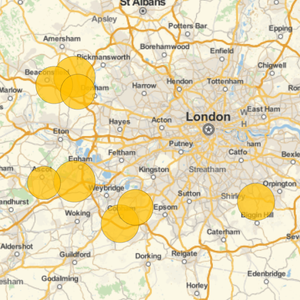
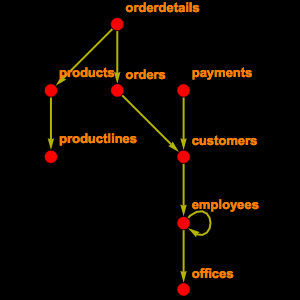
Connect to a Public MySQL Instance
In bioinformatics, there are a number of public SQL endpoints that contain very large datasets. This example shows how easy it is to connect to one and to quickly extract information that would be very difficult to process in memory.
In order to see some data, you have to connect to the public endpoint provided by the ensembl project and extract the schema information for two tables.
Construct an EntityStore.
Then you can register it.
Since you are offloading the computation to the external database, everything is very fast.
There are over 2.5 million rows in the table and you have been able to count them in a fraction of a second (including a network roundtrip).
An interesting question is, What are the most common biotypes for genes? First you are going to group by biotype and count the genes.
Then sort by the "count" property and take just the 10 largest.
Note that the last two operations are purely symbolic. To execute the query, call EntityValue.
Plot the data.